Key molecular tools employed for microbial water quality assessment
Microbial water quality assessment involves the analysis of water samples to determine the presence and concentration of microorganisms, including bacteria, viruses, and other pathogens. Various molecular tools are employed to accurately and efficiently assess microbial water quality. Some of these tools as described in Lisa Paruch her paper Int. J. Environ. Res. Public Health 2022, 19, 5128 include:
- Polymerase Chain Reaction (PCR): PCR is a widely used technique that amplifies specific DNA sequences. It allows for the detection and quantification of target microorganisms by using specific primers that match the DNA of the microorganism of interest. Real-time PCR (qPCR) provides quantitative data and is commonly used for waterborne pathogen detection.
- Quantitative Real-Time PCR (qPCR): This is an advanced version of PCR that allows for real-time monitoring of DNA amplification. It quantifies the initial amount of DNA present in a sample and is used to estimate the abundance of specific microorganisms.

- Reverse Transcription PCR (RT-PCR): RT-PCR is used to amplify RNA molecules. It is commonly applied to detect RNA viruses, such as norovirus and hepatitis A virus, which are important indicators of fecal contamination in water.
- Loop-Mediated Isothermal Amplification (LAMP): LAMP is an isothermal nucleic acid amplification technique that rapidly amplifies DNA at a constant temperature. It is particularly useful for field-based applications due to its simplicity and lack of reliance on thermal cycling equipment.
- Fluorescence In Situ Hybridization (FISH): FISH involves labeling specific DNA sequences with fluorescent probes, allowing for the visualization of microbial cells under a fluorescence microscope. It provides information on the identity and abundance of microorganisms directly in the sample.
- Microarray Technology: Microarrays are platforms that allow for the simultaneous detection of multiple target DNA or RNA sequences in a single assay. They are used to identify various microorganisms and assess their presence and abundance in water samples.
- Next-Generation Sequencing (NGS): NGS technologies, such as Illumina and Ion Torrent platforms, enable the rapid sequencing of DNA and RNA from complex microbial communities. Metagenomic and metatranscriptomic analyses provide insights into the entire microbial composition of water samples.
- High-Throughput Sequencing (HTS): HTS refers to sequencing technologies that produce a large volume of sequence data in a short period. It is particularly useful for studying diverse microbial communities in water sources.
- Digital PCR (dPCR): dPCR is a highly sensitive technique that partitions a DNA sample into numerous individual reactions. It allows for absolute quantification of target DNA molecules, making it useful for low-level pathogen detection.
- Nucleic Acid Sequence-Based Amplification (NASBA): NASBA is an isothermal amplification technique used to amplify RNA sequences. It is often applied to detect RNA viruses in water samples.
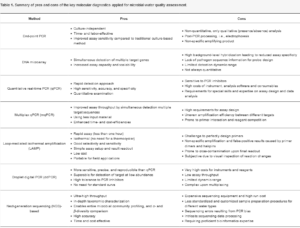
These molecular tools offer different levels of sensitivity, specificity, and throughput, allowing researchers and water quality professionals to tailor their approaches based on the specific microbial targets and the scale of assessment required.
Reference: Lisa Paruch her paper Int. J. Environ. Res. Public Health 2022, 19, 5128
More information about Key Molecular Tools employed for Microbial Water Quality Assessment? Read this paper.
Also interested in Molecular Assay Development? Contact us!


